AsianScientist (Apr. 26, 2017) – In a study published in Cell Research as a cover story, researchers from Institute of Biophysics (IBP) of Chinese Academy of Sciences have developed a new 3D genome imaging technique named TTALE that can precisely visualize specific genomic repetitive-sequence loci.
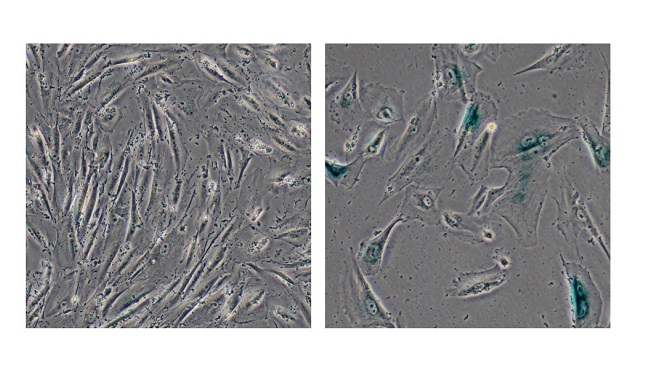
The study identified ribosomal DNA (rDNA) attrition as a hallmark of aging in three models of human mesenchymal stem cell aging, as well as in the peripheral blood cells isolated from aged human individuals.
Aging, the major risk factor for a wide range of chronic diseases, is determined by both genetic and epigenetic factors. Although telomere shortening is a hallmark of aging, repeat sequences are considered the ‘dark matter’ of the human genome and have not been studied in relation to their function in regulating biological processes including aging.
Accurate technology to track the chromatin dynamics in live cells is required for understanding the deeper mechanisms of aging process. So far, several technologies, including fluorescent in situ hybridization (FISH), TALE and CRISPR/Cas9, have been utilized to image specific loci in human genome. However, they are limited by low signal to noise ratio and transfection efficiencies, and can only be used in non-live cells.
In particular, conventional TALEs are prone to forming insoluble aggregates, hampering their utility for precise genomic imaging in human cells. To overcome this barrier, researchers screened a panel of peptides/proteins that are known to facilitate the expression of insoluble proteins in E. coli and fused them to TALE. They found that the fusion of the thioredoxin to TALE (designated as TTALE) effectively eliminated the aggregate-like signals from conventional TALE.
TTALEs were tested in a variety of cellular contexts, including human cancer cell lines, human embryonic stem cells, iPSCs and differentiated cells, as well as mouse cells in vitro or in vivo.
“TTALEs can be applied to various human cell types and mouse cells, with imaging quality comparable to 3D-FISH and transfection efficiency better than the CRISPR/dCas9 system,” said Professor Liu Guanghui from IBP.
In addition, the researchers were able to use TTALEs to precisely visualize the location of 28S rDNA inside the nucleolus of live cells, a technique which could be a powerful tool for exploring the function of rDNA and the nucleolus in different biological processes.
Although the findings in this study focused on the ability of TTALEs to explore the chromatin dynamics of aging, the application of TTALEs could be extended far beyond, such as studying single nucleotide polymorphisms-induced pathologies like autism-related disorders and detecting the chromosomal or cell cycle aberrations in cancer.
The article can be found at: Ren et al. (2017) Visualization of Aging-associated Chromatin Alterations with an Engineered TALE System.
———
Source: Chinese Academy of Sciences; Photo: Shutterstock.
Disclaimer: This article does not necessarily reflect the views of AsianScientist or its staff.